African Carnivore Connectivity Project
The African Carnivore Connectivity Project (ACCP) is a collaboration of field studies using high throughput genetic sequencing to understand connections between ecosystems and fight illegal wildlife trafficking. The ACCP began as a collaboration between Goran Spong, Matt Becker and Scott Creel but has rapidly grown by joining efforts with established field studies across Eastern and Southern Africa.
Species

The species included in the project are African wild dogs (Lycaon pictus), lions (Panthera leo), cheetahs (Acinonyx jubatus) , leopards (Panthera pardus) and spotted hyenas (Crocuta crocuta).
Locations
Geographically, the project began with the field sites of the Zambian Carnivore Programme in the Luangwa Valley Ecosystem in eastern Zambia, the Greater Kafue Ecosystem in central Zambia and the Greater Liuwa Ecosystem in western Zambia, coupled with sites in Tanzania and Mozambique from our past and
present field work. Because Zambia adjoins eight nations, it is pivotal for connectivity across eastern and southern Africa, as exemplified by the >500,000 km2 Kavango-Zambezi Transfrontier Conservation Area.
As established field studies have joined the project, the the scope has expanded to include many of the major stronghold populations of these species in ecosystems actoss eastern and southern Africa. A summary of the current scope of sampling by species and nation is in the "Collaborators" section below.
Goals
By developing collaborations with field studies of large carnivores across the region, the ACCP has begun to:
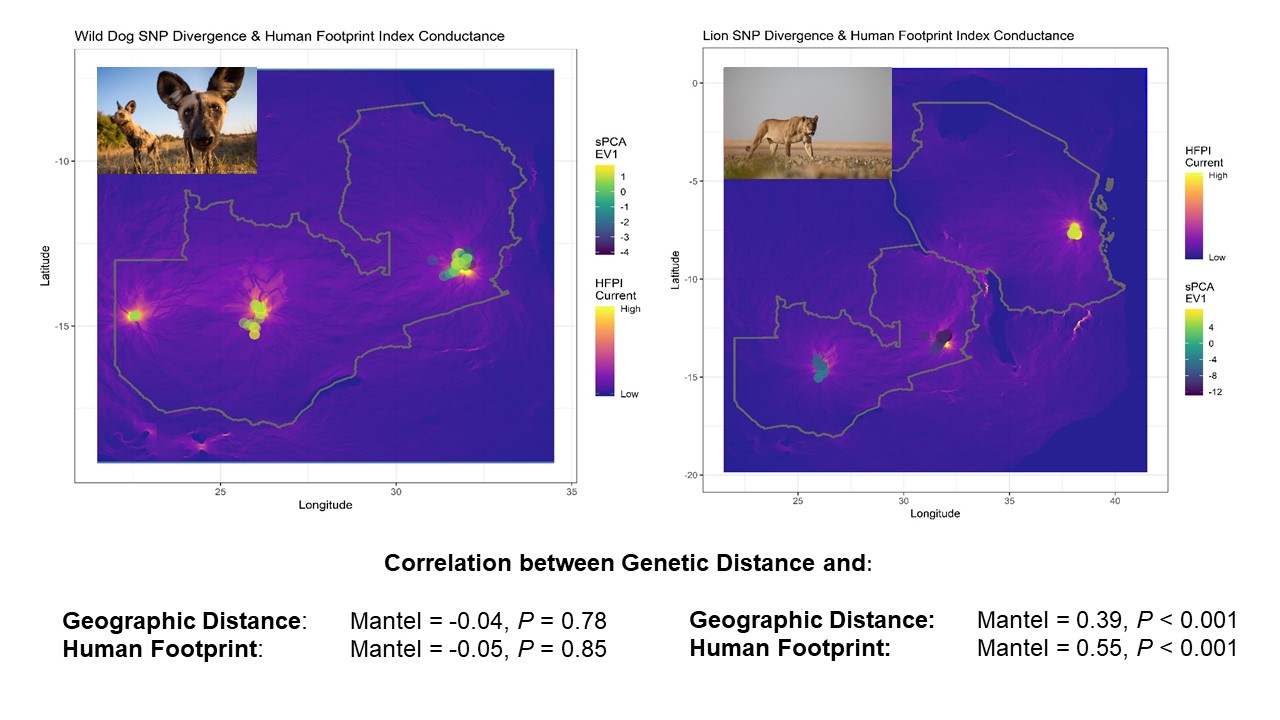
1. Quantify genetic connectivity between ecosystems, quantify differences between species, and test the factors that affect them.
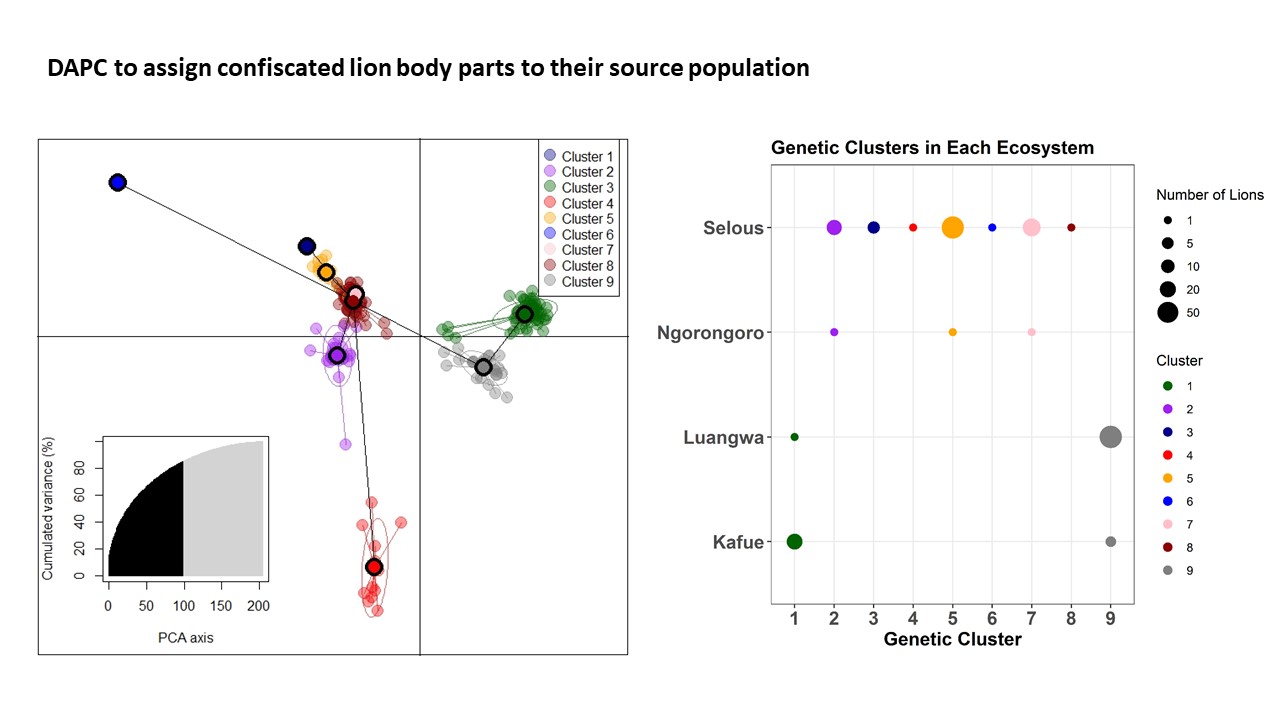
2. Build genetic profiles that allow confiscated wildlife traffic to be assigned to the ecosystem of origin.
3. Provide the data and analysis to range state governments to assist them with actions to maintain connectivity and combat illegal trafficking efficiently and effectively.
Approaches
Genotyping by Sequencing
Initally, we are using genotyping by sequencing through high throughput pyrosequencing on Illumina platforms (HiSeq 2500 and NovaSeq) at the National Genomics Insitutute in Stockholm, which allows us to identify and score genotypes at several thousand single nucleotide polymorhpism, or SNP, loci. We recently published (Scientific Reports, in press) a demonstration that this approach efficiently yields genotypes at several thousand loci for several hundred lions and wild dogs, with good power to address the goals outlined above.
This approach requires DNA extracted from high quality samples (tissue, white blood cells) . Samples from 10-20 individuals from an ecosystem are sufficient.
in parallel, we have begun developing SNP chips for the Fluidigm Biomark platform, directly run in Spong's lab at SLU. This approach provides SNP genotypes at fewer loci, but at lower cost and allowing DNA from lower quality samples (hair, feces) to be analyzed. Our prior work with brown bears and woverines shows that these data provide plenty of power to address the goals above.
Statistical Analysis
We are using a combination of t-distributed stochastic neighbor embedding (van der Maaten 2014) and spatial PCA (Jombart et al. 2008) to quantify spatial patterns of genetic similarity and difference between ecosystems. We are using disciminant analysis of principal components (Jombart et al 2010) to assign genotypes from confiscated traffic to the ecosystem of origin.
Other Output
The SNP genotypes described above have enough statistical power to address questions about genetic patterns within ecosystems. For example, they clearly distinguish between first-order (r=0.5, e.g. parent-offspring), second-order (r=0.25, e.g. half siblings) and unrelated pairs of individuals (shown below for several thousand pairs of lion genotypes). The SNP genotypes from each project's samples will be returned to each project (in the 0/1/2 gds format used by the R/Bioconductor package SNPRelate) for within-ecosystem analyses.
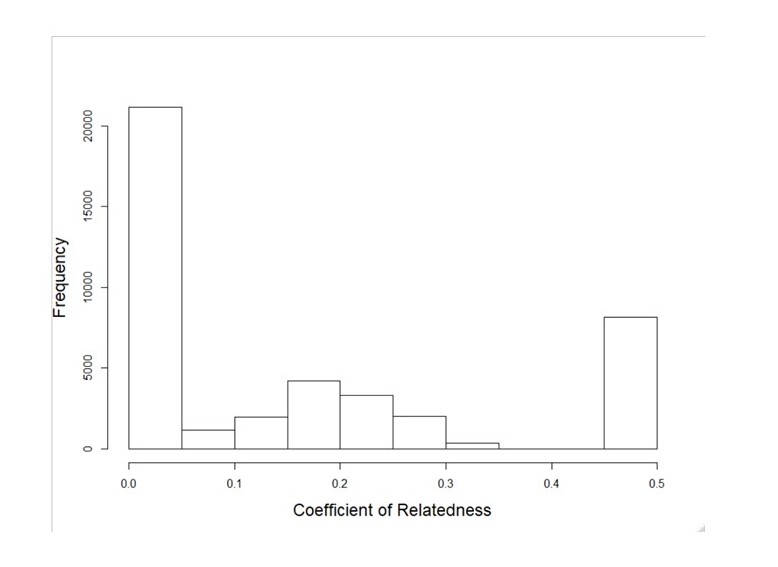 With sufficient sampling within a population, these data allow reconstruction of pedigrees,
enabling studies of mating patterns, reproductive success and contemporary dispersal.
With sufficient sampling within a population, these data allow reconstruction of pedigrees,
enabling studies of mating patterns, reproductive success and contemporary dispersal.
Collaborators
Collectively, we have a long history of informal collaboration and we are comfortable working with like-minded field studies who (A) have a use for genotypes from their study animals and (B) would like to participate in broader research on connectivity between ecosystems and/or contribute to a baseline database to better understand the source of illegal traffic in large carnivore body parts. Contact any of the PIs: Creel, Becker, Spong.
Current Collaborators
Collaborators in ACCP include: Sarah Durant (Zoological Society of London), Rosemary Groom (Zoological Society of London), Chuma Simukonda (Zambia DNPW), Andrew Loveridge (Oxford WildCRU), David Macdonald (Oxford WildCRU), Simon Dures (Edinburgh, TRACE), Paola Bouley (Gorongosa), Colleen Begg (Niassa), Faruk Mamugy (Mozambique), Valerio Macandza (Mozambique), Emma Stone (Bristol), Ingela Jansson (Ngorongoro, SLU), Kerri Rademeyer (Wildlife Crime Prevention) and Rob Ogden (Edinburgh, TRACE)
Scope of Sampling
The current scope of the project, by species and nation:

Publications
Creel S, Spong G, Becker M, Simukonda C, Norman A, Schiffthaler B, Chifunte C. 2019. Carnivores, competition and genetic connectivity in the Anthropocence. Nature Scientific Reports, 9, 16339 (2019) doi:10.1038/s41598-019-52904-0



